點(diǎn)擊化學(xué)Protocols
使用熒光疊氮化物對炔烴標(biāo)記的DNA進(jìn)行成像Protocol
EdU Imaging, Microscopy ![]() 下載為PDF
下載為PDF
This protocol was tested with A549, HeLa, and NIH/3T3 cells but it can be adapted for any adherent cell type. In initial experiments, we recommend starting with 5 μM concentration of an azide detection reagent. Slight adjustment of an azide detection reagent concentration might be required in case of high background or low signal intensity.
Fluorescent azides, and EdU can be ordered from Click Chemistry Tools.
Prepare the following reagents:
? 100 mM copper sulfate in water
? 100 mM sodium ascorbate in water (20 mg/mL, should be used freshly prepared)
? 2 mM (ca. 2 mg/mL) azide labeling reagent in water or DMSO
? Fixative (for example 3.7% Formaldehyde in PBS)
? Permeabilization reagent (for example, 0.5% solution of Triton? X-100 in PBS)
? 3% BSA in PBS (pH 7.4)
? Coverslips/microscope slides, mounting media
? PBS buffer, pH 7.4
? Deionized water
Cell fixation and permeabilization
The following protocol is provided for fixation step using 3.7% formaldehyde in PBS followed by a 0.5% Triton?X-100 permeabilization step. Protocols using other fixation/permeabilization reagents, such as methanol and saponin also can be used.
1.1 Transfer each coverslip into a single well. For convenient processing use 6-well plates.
1.2 After EdU labeling, remove the media and add 1 mL of 3.7% formaldehyde in PBS to each well containing the coverslips. Incubate for 15 minutes at room temperature.
1.3 Remove the fixative and wash the cells in each well twice with 1 mL of 3% BSA in PBS.
1.4 Remove the wash solution. Add 1 mL of 0.5% Triton? X-100 in PBS to each well, then incubate at room temperature for 20 minutes.
EdU detection
Note: 500 μL of the reaction cocktail is used per coverslip. A smaller volume can be used as long as the remaining reaction components are maintained at the same ratios.
2.1 Prepare required amount of the reaction cocktail according to Table 1. Add the ingredients in the order listed in the table. Use the reaction cocktail within 15 minutes of preparation.
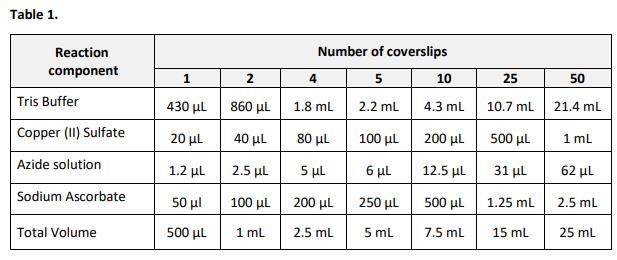
2.2 Remove the permeabilization buffer (step 1.4). Wash the cells in each well twice with 1 mL of 3% BSA in PBS. Remove the wash solution.
2.3 Add 0.5 mL of the Reaction Cocktail to each well containing a coverslip. Rock the plate briefly to insure that the reaction cocktail is distributed evenly over the coverslip.
2.4 Protect from light, and incubate the plate for 30 minutes at room temperature.
2.5 Remove the reaction cocktail.
Wash each well once with 1 mL of 3% BSA in PBS.
Remove the wash solution.
At this point the samples are ready DNA staining. If no DNA staining is desired, proceed to Imaging.
If antibody labeling of the samples is desired, proceed to labeling according to manufacturer’s recommendations. Keep the samples protected from light during incubation.
DNA staining with Hoechst 33342 (Optional)
3.1 Wash each well with 1 mL of PBS. Remove the wash solution.
3.2 Prepare 1 x Hoechst 33342 solution (5 μg/mL.) Final concentrations of 1x Hoechst 33342 may range from 2 μg/mL to 10 μg/mL.
3.3 Add 1 mL of 1x Hoechst 33342 solution per well. Protected from light. Incubate for 30 minutes at room temperature.
3.4 Remove the Hoechst 33342 solution.
3.5 Wash each well twice with 1 mL of PBS.
3.6 Remove the wash solution.
